2007
Journal Publication
Genome Research
MAKER: An Easy-to-use Annotation Pipeline Designed for Emerging Model Organism Genomes
Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M
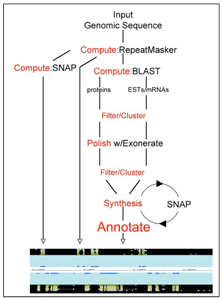
We have developed a portable and easily configurable genome annotation pipeline called MAKER. Its purpose is to allow investigators to independently annotate eukaryotic genomes and create genome databases. MAKER identifies repeats, aligns ESTs and proteins to a genome, produces ab-initio gene predictions, and automatically synthesizes these data into gene annotations having evidence-based quality indices. MAKER is also easily trainable: outputs of preliminary runs are used to automatically retrain its gene-prediction algorithm, producing higher quality gene-models on subsequent runs. MAKER’s inputs are minimal and its outputs can be used to create a GMOD database. Its outputs can also be viewed in the Apollo Genome browser; this feature of MAKER provides an easy means to annotate, view, and edit individual contigs and BACs without the overhead of a database. As proof of principle we have used MAKER to annotate the genome of the planarian Schmidtea mediterranea and to create a new genome database, SmedGD. We have also compared MAKER’s performance to other published annotation pipelines. Our results demonstrate that MAKER provides a simple and effective means to convert a genome sequence into a community accessible genome database. MAKER should prove especially useful for emerging model organism genome projects for which extensive bioinformatics resources may not be readily available.
Address reprint requests to: Alejandro Sánchez Alvarado